Projects
Predicting Drug Dose Responses Using GNNs and Attention Mechanisms
Combination therapies have emerged as a powerful treatment modality to overcome drug resistance and improve treatment efficacy. However, the rapid increase of drug combinations with the number of individual drugs in consideration has led to in Silico simulations becoming widespread. While many studies are only able to find synergistic pairs of drugs, our method which is named DeepDDR obtains the full dose–response matrices of different drug tuples which enables finding the optimal dose required for many applications. Additionally, this approach offers a more in-depth view of the drugs’ complex response landscape, and allows to calculate different synergy metrics as a follow-up step.
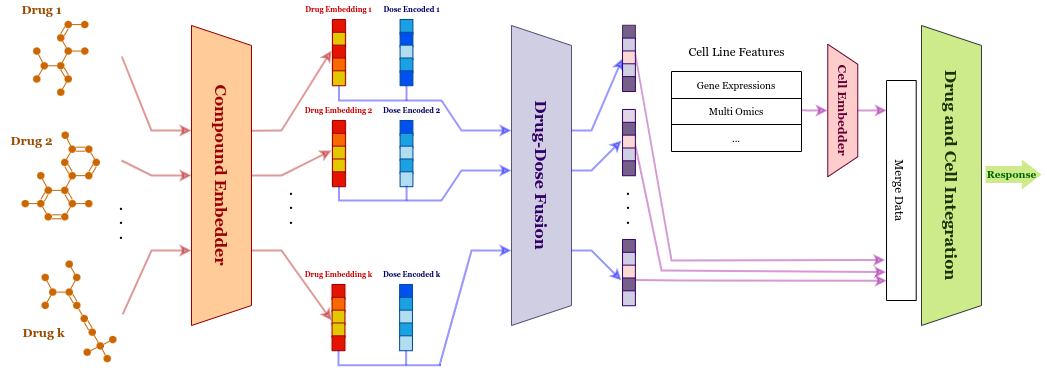
From a technical perspective, DeepDDR facilitates Graph Neural Networks and multi-head attention with the goal of identifying the most effective drug combinations for further pre-clinical and clinincal validation. In this work, we propose a general recipe for devising Deep Neural Network architectures that can predict dose responses for multiple drugs on a specific cell-line. In our opinion, this framework can inspire future work to create new models and further address the drug-dose response problem. Graph Neural Networks and Attention-based models have emerged as powerful machine learning tools in many applications; therefore, we incorporate popular architectures in the domain to obtain latent embeddings of drug compounds and combine cell-line features with these embeddings. That being said, while previous work on drug-dose response considers MACCS fingerprints to represent drug compounds, we follow the trend of using Graph Neural Networks on SMILES notations of compounds to produce more accurate embeddings of drugs. These embeddings can take into account the structure of the compound as a whole. Finally, our findings indicate that using DeepDDR yields state-of-the art results on the comprehensive drug-dose-response NCI-ALMANAC dataset which contains drug pair dose responses on popular human cancer cell-lines.
Hamidreza Kamkari, Karim Abbasi
Semidefinite Programming using a Physarum Based Dynamic
 |
Physarum Polycephalum is a slime mold that can solve shortest path problems. A mathematical model based on Physarum's behavior, known as the Physarum Directed Dynamics, can solve positive linear programs. In this paper, we present a family of Physarum-based dynamics extending the previous work and introduce a new algorithm to solve positive Semi-Definite Programs (SDP). The Physarum dynamics are governed by orthogonal projections (w.r.t. time-dependent scalar products) on the affine subspace defined by the linear constraints. We present a natural generalization of the scalar products used in the LP case to the matrix space for SDPs, which boils down to the linear case when all matrices in the SDP are diagonal, thus, representing an LP. |
We investigate the behavior of the induced dynamics theoretically and experimentally, highlight challenges arising from the non-commutative nature of matrix products, and prove soundness and convergence under mild conditions. Moreover, we consider a more abstract view of the dynamics that suggests a slight variation to guarantee unconditional soundness and convergence-to-optimality. By simulating these dynamics using suitable discretizations, one obtains numerical algorithms for solving positive SDPs, which have applications in discrete optimization, e.g., for computing the Goemans-Williamson approximation for MaxCut or the Lovasz theta number for determining the clique/chromatic number in perfect graphs.
Yuan Gao, Hamidreza Kamkari, Andreas Karrenbauer, Kurt Mehlhorn, Mohammadamin Sharifi (PDF) (Code)
ML-Mnemonist
 |
This is an indie self-developed project to simplify the ML operations needed for developing an ML model. The framework currently only focuses on the developmental stage of ML operations and is a lightweight package to create ML experiments, store and manage experiment configurations, hyperparameter tuning. One unique aspect of this project is its caching capabilities. Caching is done at the software level and one can use this package to work with any remote server. For example, by combining ML-Mnemonist with a Google Colab environment, you do not need to worry anymore about losing your progress when the session crashes or gets disconnected for any reason. |
Dental Panoramic Image Reconstruction and Tooth Segmentation
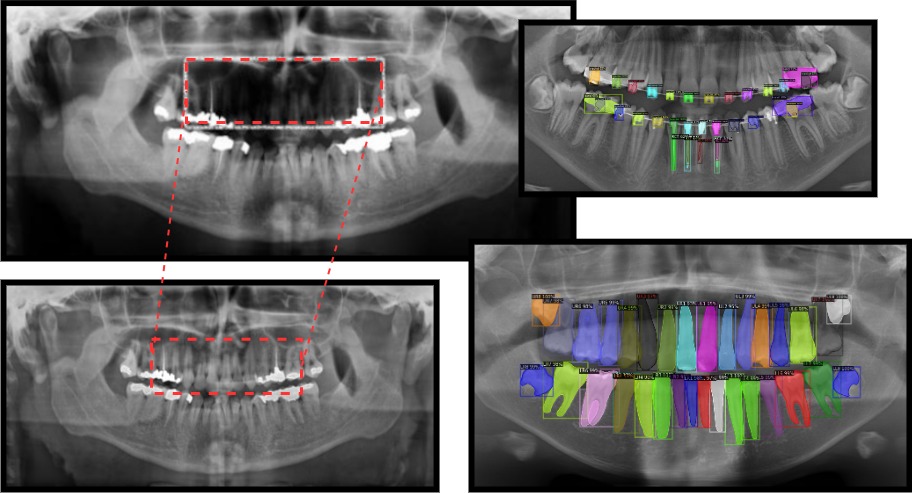
Computer-aided diagnosis has become widespread in dental applications and with the advent of data-driven approaches in dealing with radiographic images, machine learning models are being deployed in order to help dentists in the visual representation of images as well as aiding them in their diagnosis process. During my experience with Fanap, I experimented with U-Net type architectures for panoramic dental image correction and tooth segmentation based upon Mask-RCNN type architectures. Our results, demonstrate our capabilities in restoring super exposed dental images and also the ability to accurately segment teeth and dental augmentations performed on the patient.
Fine-Grained Complexity of Optimizing Bias Terms in Neural Networks
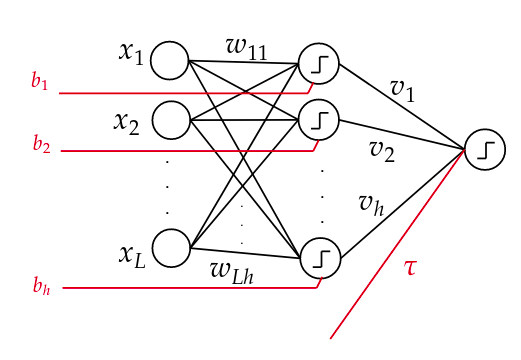 |
The study of the complexity of learning neural networks is of high importance in the infinite quest of shedding light upon some theoretical aspects of them. One particular problem of interest is the problem of fine-tuning such networks. To explore the problem, we limit ourselves to a subset of weights and try to fix the other weights while optimizing this set according to the input data. In this work, we have addressed the complexity of fine-tuning bias terms in a fine-grained fashion and proved that even for a simple one-hidden layer neural network we require a lot of processing time to optimize these weights deterministically. |
Andreas Karrenbauer, Hamidreza Kamkari, Karl Bringmann
RNA Sequence Design using Graph Neural Networks
RNA is one of the major classes of information-carrying biopolymers in the cell of living organisms and recent studies revealed a key role in regulatory processes and transcription control, which have also been connected to certain diseases. Therefore, the engineering of RNA molecules is of growing importance with applications ranging from biotechnology and medicine to synthetic biology. The functional properties of RNA are derived from their secondary structure. Given a secondary structure, we aim to predict sequences that fold into a target minimum free energy secondary structure, while considering various constraints. This problem, known as RNA Inverse Folding or RNA Sequence Design, can be regarded as a combinatorial optimization problem and in our work, we aim to solve this using a data-driven approach.
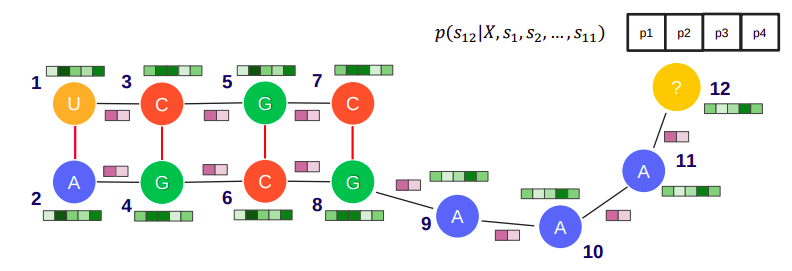
Specifically, we present a Graph Neural Network (GNN) based model that learns an autoregressive decomposition of the distribution of nucleotides given a certain secondary structure. The structure is embedded using our GNN and by carefully sampling from this generative model we can obtain the correct solution to the RNA inverse folding problem. Our model, while still improving, has proven to yield good results on RNA inverse folding datasets such as PseudoBase+++, Rfam-Tenda, and Eterna100.
Mehdi Saman Booy, Hamidreza Kamkari, Alexander Ilin, Pekka Orponen